Housheng Hansen He 博士/高级研究员,副教授(玛格丽特公主癌症中心,加拿大多伦多大学)
报告题目:The transcriptomic and functional landscape of circRNAs in cancer

嘉宾简介:
2003年获得北京师范大学物理学学士学位,之后转入基因组学和非编码RNA研究领域,2008年获得中国科学院生物物理研究所博士学位。随后在哈佛大学Dana-Farber癌症研究所开展癌症基因组学和表观基因组学的博士后工作。2011年开始在哈佛大学医学院担任讲师。2013年加入玛格丽特公主癌症中心和多伦多大学,任研究员和助理教授。2018年被提升为高级研究员和副教授。Housheng Hansen He教授在高影响力的SCI学术期刊上发表了约50篇文章。2019年初,在Cell杂志发表重磅文章,系统分析了局限性前列腺癌中circRNA表达特征。
代表性成果介绍:
Cell:前列腺癌功能性circRNA筛选与分析
2019年2月7日,Cell杂志以Article形式发表了加拿大多伦多大学Paul C. Boutros教授和Housheng Hansen He教授为共同通讯作者的研究工作(参考文献[1])。本项工作共鉴定到76311个circRNA分子,还包括62个融合基因来源的circRNA分子。有趣的是,作者统计发现circRNA和对应linear RNA产物的表达丰度相关性并不高,仅有3%的circRNA表达丰度与对应的linear RNA产物丰度高度相关。绝大部分circRNA的丰度低于对应的linear RNA产物,但有127种circRNA的丰度高于对应linear RNA产物。为找出对前列腺癌有必要功能(essential function)的circRNA分子,作者设计了shRNA文库进行高通量筛选。本文设计的shRNA文库分析了1507种circRNA及它们所对应的1075个host gene线性产物的shRNA,每个RNA分子至少设计2个target位点,文库感染了四种前列腺癌细胞株。shRNA文库筛选结果发现了171种对前列腺癌细胞增殖有必要功能的circRNA分子,但它们所对应的linear RNA分子绝大部分没有必要功能(90%),相关结果也借助其他实验进行了验证分析。为进一步证明这些具有必要功能的circRNA分子的功能,作者挑选了circCSNK1G3进行分析,依据主要包括:circCSNK1G3在四种前列腺癌细胞中均具有essential function,它的线性基因则没有。进一步作者发现circCSNK1G3可竞争性结合miR-181b/d (参考文献[1])。
推荐阅读:重磅!Cell杂志同期发表两篇circRNA研究文章
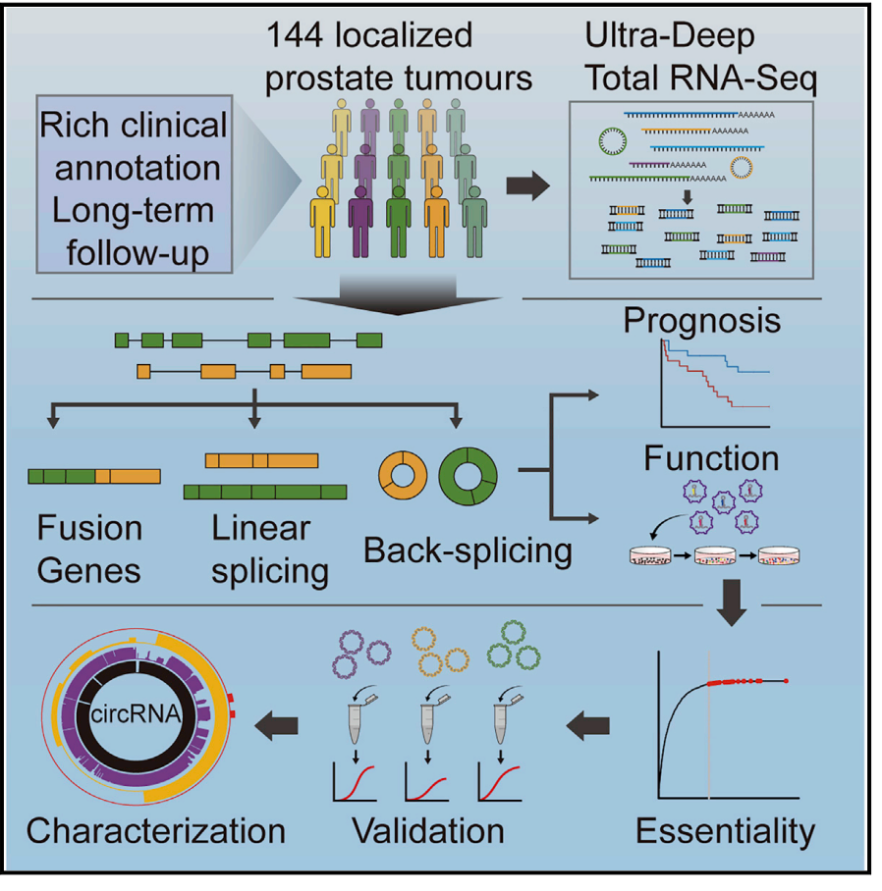
前列腺癌功能性circRNA筛选分析 (参考文献[1])
Cell:前列腺癌风险性SNP位点调控启动子-增强子转换调控lncRNA剪切型表达状态
2018年7月26日,Cell杂志发表了一项重要研究成果,报道发现前列腺癌风险相关SNP位点rs11672691通过调控启动子-增强子转换影响lncRNA PCAT19的剪切型表达状态,最终影响前列腺癌临床预后。文章的通讯作者是Housheng Hansen He教授。(参考文献[3])。
研究表明前列腺癌(PCa)中存在一百多种疾病相关的SNP位点,其中rs11672691与患者的侵袭性呈现正相关。序列分析表明rs11672691位点位于一种非编码RNA PCAT19的基因区,恰好位于短型剪切形式(PCAT19-short)的启动子区,该位点则位于长型剪切形式(PCAT19-long)的第三内含子中。s11672691位点的GG型临床预后明显比AG+AA型的病人差。数量性状遗传(eQTL)分析表明该位点与lncRNA PCAT19相关性最高(来源于471例健康前列腺组织的数据),进一步,作者发现GG型,AG型和AA型对应的PCAT19-short和PCAT19-long相对表达量有显著区别,GG型倾向于促进PCAT19-long的表达。而AG型和AA型倾向于PCAT19-short型表达。序列分析表明rs11672691位点具有启动子和增强子双重功能。ChIP结果表明转录因子NKX3.1和YY1可以优先结合非疾病相关的rs11672691等位位点,但rs11672691位点的连锁不平衡SNP rs887391的存在则降低了上述两种转录因子的结合能力,反而增强了该位点增强子的活性,导致PCAT19-long的激活和表达。PCAT19-long与HNRNPAB相互作用以激活与PCa进展相关的下游细胞周期基因,从而促进PCa肿瘤生长和转移。(参考文献[3])
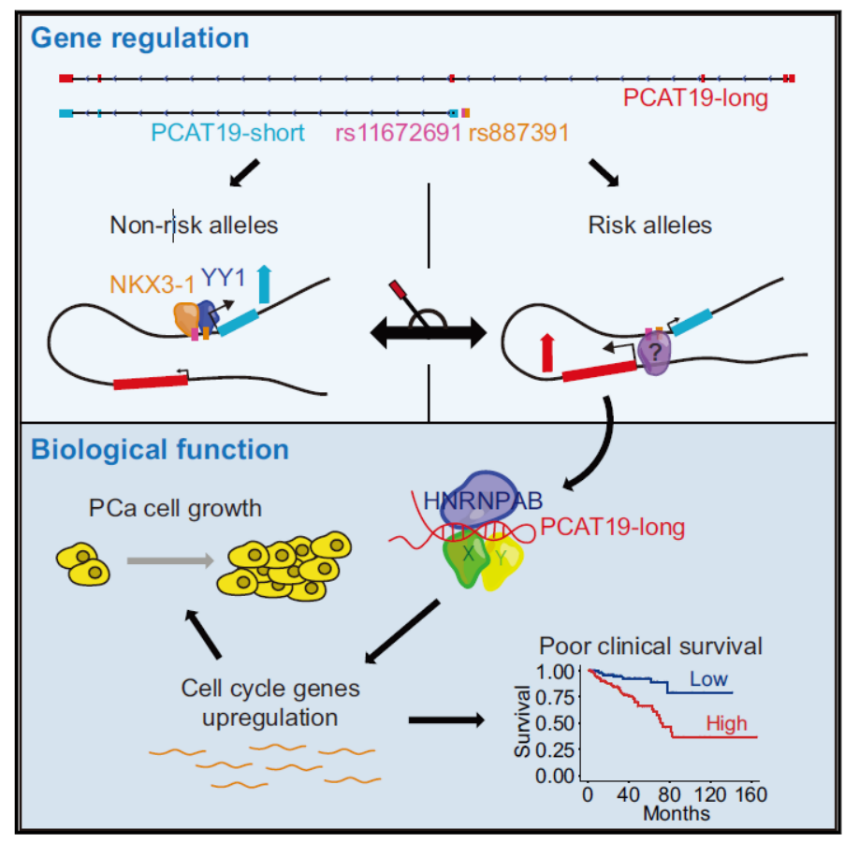
rs11672691位点不同基因型导致该位点启动子-增强子转换 (参考文献[3])
Cancer Research:前列腺癌中LSD1介导表观遗传改变调控CENPE表达
2017年10月15日,Cancer Research杂志发表了Housheng Hansen He教授为通讯作者的研究论文,报道染色质修饰相关因子LSD1在去势抵抗的前列腺癌(CRPC)中介导表观遗传重编程,调控包括CENPE在内的细胞周期相关因子的表达。LSD1是雄激素受体AR转录活性的重要调控因子。在CRPC中LSD1介导的表观遗传重编程导致一些细胞周期相关的基因表达改变,其中包括参与中心体中的一种有丝分裂的驱动蛋白CENPE。LSD1与AR一起结合CENPE的启动子,激活该基因的表达。药物抑制CENPE可显著抑制CRPC细胞增殖。(参考文献[7])
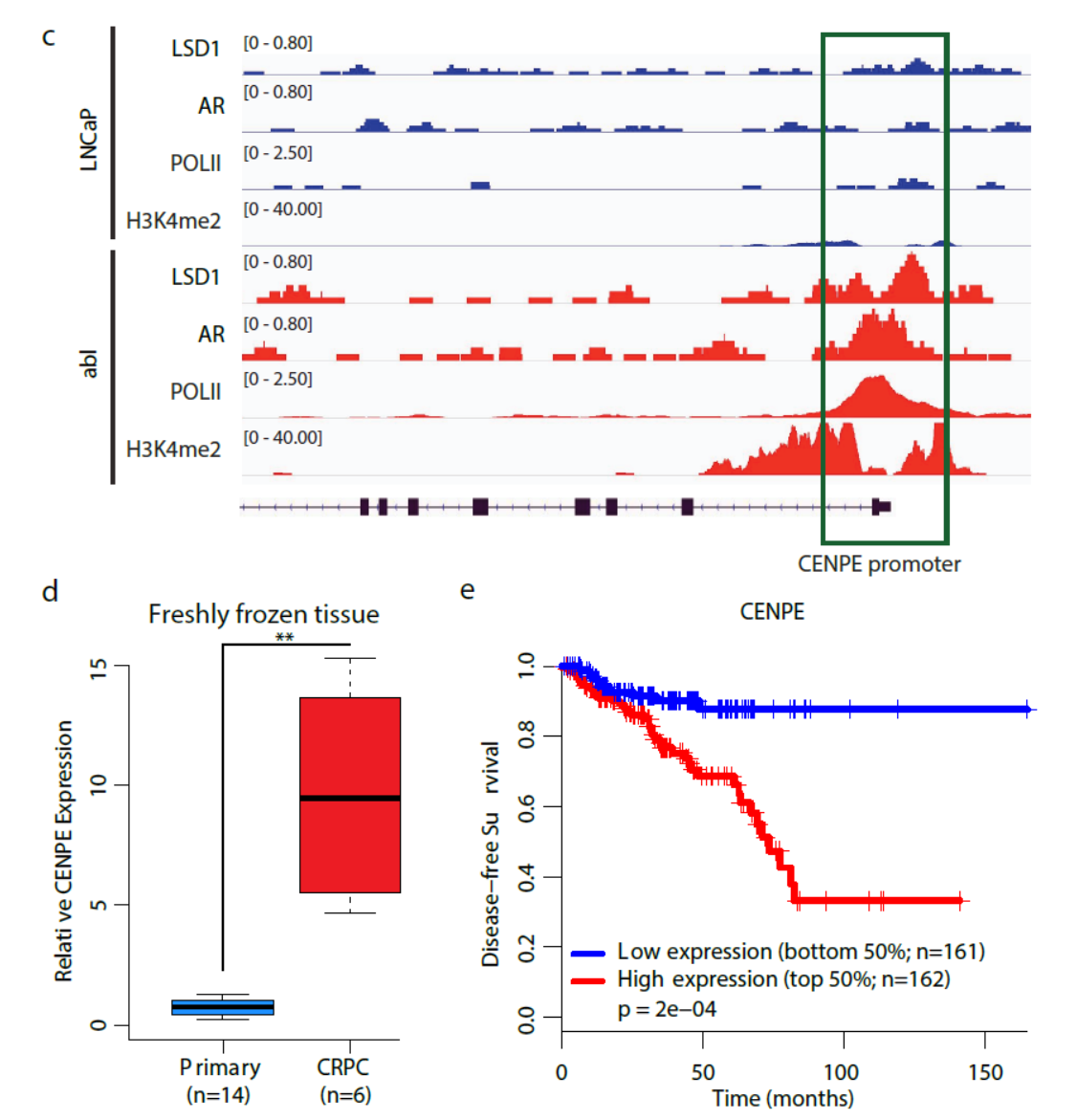
LSD1与AR共同调控CENPE表达 (参考文献[7])
PNAS:细胞周期相关转录特分析
2017年3月28日,PNAS杂志发表了一项细胞周期相关转录特征分析的文章,文章通讯作者是Dana-Farber 癌症中心的Myles Brown教授,X. Shirley Liu教授和玛格丽特公主癌症中心Housheng Hansen He教授。本文作者在MCF-7细胞中通过global nuclear run-on sequencing (GRO-seq),RNA sequencing (RNA-seq)和组蛋白修饰ChIP (ChIP-seq)分析了G0/G1, G1/S和 M期细胞中基因转录的特征。(参考文献[9])
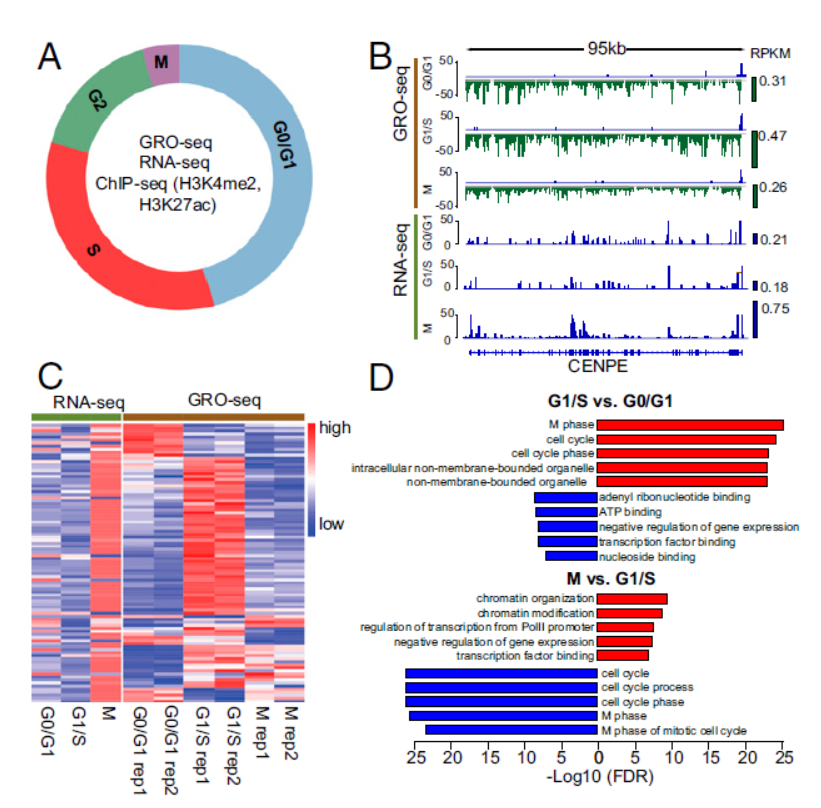
GRO-seq和RNA-seq分析细胞周期相关转录特征(参考文献[9])
Nature Genetics:前列腺癌风险相关SNP位点介导lncRNA PCAT1表达及功能研究
2016年10月,Nature Genetics杂志发表了Housheng Hansen He教授为通讯作者的文章,介绍发现前列腺癌相关SNP位点rs7463708通过促进ONECUT2和AR结合,形成增强子结构促进非编码RNA PCAT1的表达,PCAT1可通过调控LSD1和AR的结合,调控下游基因的表达,包括GNMT和DHCR24等等。(参考文献[16])
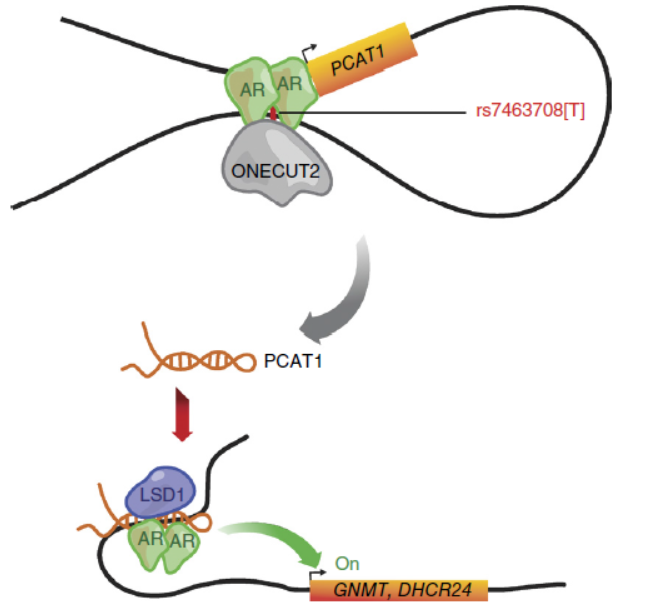
rs7463708调控PCAT1的表达(参考文献[16])
Nature:三阴性乳腺癌对BET溴结构域抑制剂的反应和耐药性机制
2016年1月21日,Nature杂志发表了Dana-Farber癌症中心James E. Bradner教授和Kornelia Polyak教授为共同通讯作者的文章,介绍三阴性乳腺癌中BET溴结构域抑制剂药物作用和耐药机制的研究。Housheng Hansen He为本文的共同第一作者。
三阴性乳腺癌因为缺乏ER等标志物,导致没有有效的靶向治疗药物,在本文之前BET溴结构域抑制剂在其他类型肿瘤中有一定效果,但在三阴性乳腺癌中这类药物的效果如何还不清楚。BET溴结构域抑制剂通过竞争性结合并抑制BET结构域蛋白(如BRD4),抑制这些蛋白与染色质的结合,从而抑制相关下游基因的表达。三阴性乳腺癌细胞系对这类抑制剂有一定的敏感性。作者进一步分析了对这类药物敏感性的细胞在诱导耐药前后的基因突变和基因表达特征的分析,结果没有发现与耐药有关的特殊基因突变和耐药基因(如一些药泵基因)的表达,并且在这类药物耐药的细胞中依然有野生型BRD4的表达,说明三阴性乳腺癌中存在其他不依赖于BET结构域蛋白的下游基因激活机制。蛋白质组学分析表明MED1,高度磷酸化的BRD4是PP2A活性下降的重要因素。(参考文献[19])
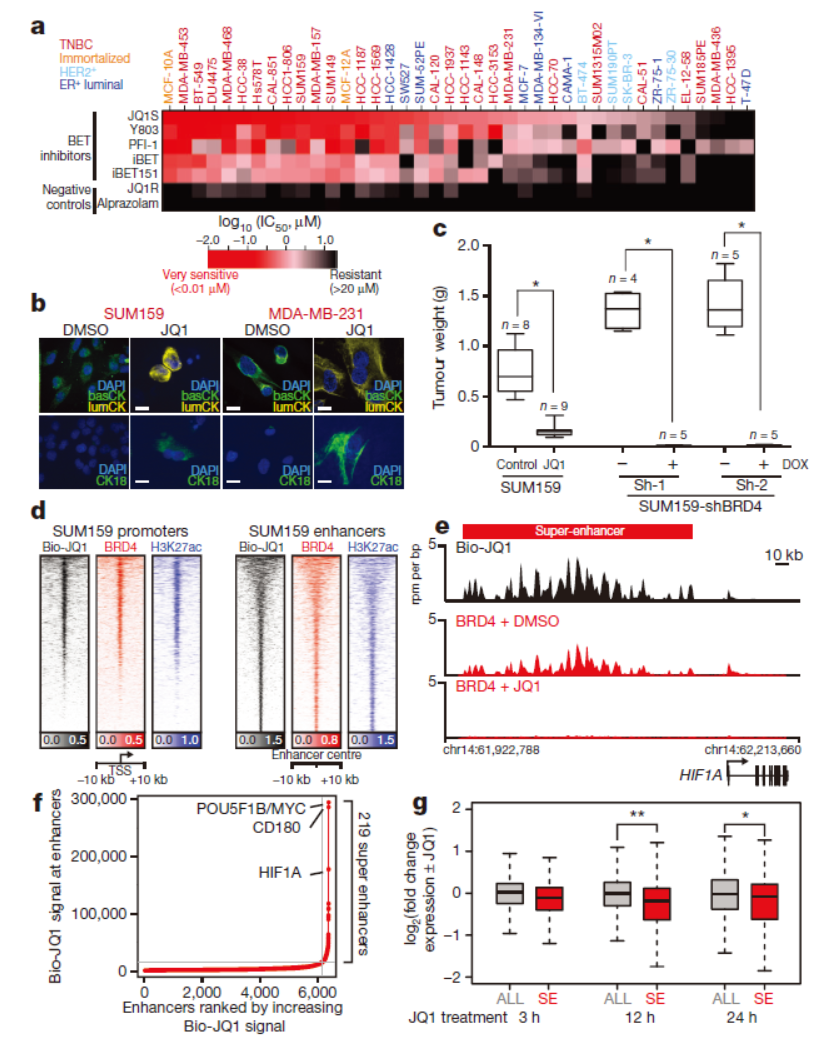
三阴性乳腺癌中BET结构域蛋白抑制剂药效分析(参考文献[19])
Nature Methods:改良的DNase-seq表明转录因子足迹分析的内在偏差
2014年1月,Nature Methods杂志发表了一项关于DNase-seq技术改良的方法学文章,通讯作者是Dana-Farber癌症中心的X Shirley Liu教授和Myles Brown教授,Housheng Hansen He为本文的共同第一作者。(参考文献[23])

Nature Genetics:核小体动力学定义增强子
2010年4月,Nature Genetics杂志发表了一篇关于增强子研究的重要研究论文,表明核小体的动力学分析有助于定义基因的增强子。文章的通讯作者是Dana-Farber癌症中心的X Shirley Liu教授和Myles Brown教授,Housheng Hansen He为本文的共同第一作者。(参考文献[37])
文中作者基于高通量测序,分析了全基因组中表观修饰的核小体动态变化情况。在前列腺癌细胞中经过雄激素处理后,发现AR结合区(通常为增强子)的一对核心核小体发生脱离。基于这一特征,作者构建了定量分析增强子的模型,并准确预测到AR和FOXA1结合的区域。作者还基于这一模型成功发现了此前未报道的长期雄激素处理后OCT1和NKX3-1的结合位点特征。本文所构建的增强子结构的定量建模提供了一种强有力的预测方法,可用于预测参与细胞对特定刺激的反应的转录因子。(参考文献[37])
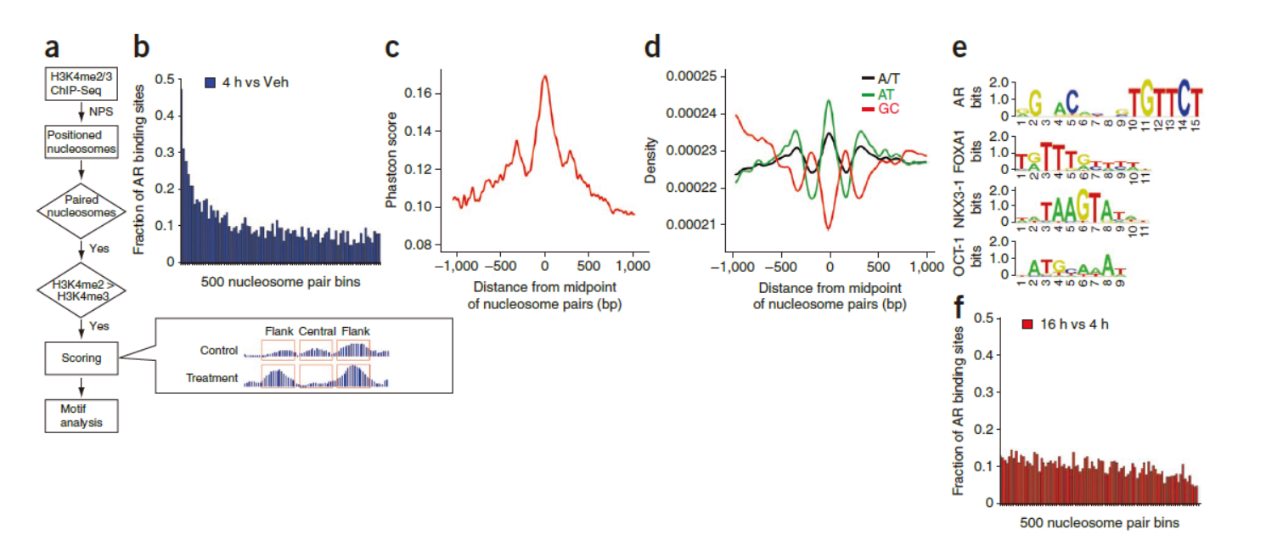
成对核小体变化的定量模型 (参考文献[37])
Housheng Hansen He教授实验室主页:http://hansenhelab.org/
已发表相关文献:
1. Chen S., … ,Boutros PC, He HH.#. Widespread and Functional RNA Circularization in Localized Prostate Cancer. 2019, Cell
2. Quigley DA., …, He HH. , …, Feng F. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. 2018, Cell
3. Hua JT., …, He HH.#. Risk SNP-Mediated Promoter-Enhancer Switching Drives Prostate Cancer through lncRNA PCAT19. 2018. Cell
4. Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, Li Y, Chen H, Yang H, Hsu PH, Van Allen EM, Freeman GJ, De Carvalho DD, He HH, Sharpe AH, Shi Y. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. 2018. Cell. PMID: 29937226.
5. Ci X, Hao J, Dong X, Choi S, Xue H, Wu R, Qu S, Gout PW, Fang Zhang F, Haegert AM, Fazli L, Crea F, Ong C, Zoubeidi A, He H, Gleave M, Collins C, Lin D, Wang YZ. A heterochromatin gene signature unveils HP1α mediating neuroendocrine prostate cancer development and aggressiveness. 2018. Cancer Research. PMID: 29487201.
6. Han W*, Gao S*, Barrett D, Han D, Macoska J, Ahemd M, He H.#, Cai C#. Reactivation of Androgen Receptor-Regulated Lipid Biosynthesis Drives the Progression of Castration-Resistant Prostate Cancer. 2018. Oncogene. PMID: 29059155.
7. Liang Y*, Ahmed M*, Guo H, Soares F, Hua J, Gao S, Lu C, Poon C, Langstein J, Ekram M, Li B, Davicioni E, Takhar M, Erho N, Karnes R, Chadwick D, Kwast T, Boutors P, Arrowsmith C, Feng FY, Joshua A, Cai C, He H.#. LSD1 Mediated Epigenetic Reprogramming Drives CENPE Expression and Prostate Cancer Progression. 2017. Cancer Research. PMID: 28916652.
8. Fei T*, Chen Y*, Xiao T, Li W, Cato L, Zhang P, Cotter MB, Bowden M, Rosina LT, Zhao SG, Wu Q, Feng Y, Loda M, He H, Liu X, Brown M. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. 2017. PNAS. PMID: 28611215.
9. Liu Y*, Chen SJ*, Wang S, Soares F, Fischer M, Decaprio JA, Meyer C, Brown M#, Liu X#, He H#. Transcriptional Landscape of the Breast Cancer Cell Cycle. 2017. PNAS. PMID: 28289232.
10. Ahmed M, Sallari R, Guo H, Moore J, He H#, Lupien M#. Variant Set Enrichment: A Fast Method to Identify Disease-Associated Functional Genomic Regions. BioData Mining. 2017. PMID: 28239419.
11. Hua J, Lu J, Isaev K, Ahmed M, Guo H, Soares F, He H#. Noncoding RNA for personalized prostate cancer treatment: utilize the “dark matters” of the genome? Personalized Medicine. 2017.
12. Guo H*, Ahmed M*, Hua J, Soares F, He H#. Crucial role of noncoding RNA in driving prostate cancer development and progression. Epigenomics. 2017. PMID: 27885842.
13. Hao ZY, Sheng Y, Li WY, Duncan GS, Sylvester J, Lin GHY, Snow BE, Brenner D, You-Ten A, Su YW, Haight J, Inoue S, Wakeham A, Elford A, Hamilton S, Liang Y, Zuniga-Pflucker JC, He H, Ohashi PS, Mak TW. K48-linked ubiquitination of KLF4 by the E3 ligase Mule controls T cell proliferation through cell cycle regulation. Nature Communications. 2017. PMID: 28084302.
14. Fraser M, Sabelnykov V.Y, Yamaguchi T.N, Heisler L.E, Livingstone J, Huang W,… He H, Fradet Y, Tetu B, Kwast T, McPherson J.D, Bristow R.G, Boutros P.C. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2016. PMID: 28068672.
15. Wang S, Zang C, Xiao T, Fan J, Mei S, Qian Q, Wu Q, Xu K, He H, Brown M, Meyer CA, Liu X.S. Modeling cis-regulation with a compendium of genome-wide histone H3K27ac profiles. Genome Research. 2016. PMID: 2746632.
16. Guo H*, Ahmed M*, Liang Y, Hua J, Langstein J, Poon C, Bailey S, Desai K, Fei T, Li Q, Prensner JR, Pomerantz M, Feng FY, Freedman M, Lupien M, He H#. Modulation of long noncoding RNAs underlying genetic predispositions to prostate cancer. Nature Genetics. 2016. PMID: 27526323.
17. Zang C, Wang T, Deng K , Li B, Hu S, Qin Q, Xiao T, Zhang S, Meyer CA, He H, Brown M, Liu J, Xie Y, Liu X. High-dimensional genomic data bias correction and data integration using MANCIE. Nature communications. 2016. PMID: 27072482.
18. Ma F, Ye H, He H, Gerrin SJ, Chen S, Tanenbaum BA, Cai C, Sowalsky A, He L, Wang H, Balk SP, Yuan X. SOX9 drives WNT pathway activation in Prostate Cancer. J Clin Invest. 2016. PMID: 27043282.
19. Shu S*, Lin CY*, He H*, Doherty E*, Brown J, Mohammed H, D’Santos C, Mckeown M, Ott C, Qi J, Ni M, Rao PK, Duarte M, Chiang C, Anders L, Young RA, Carroll JS, Long H, Brown M, Liu XS, Meyer CA, Bradner JE, Polyak K. Response and resistance to BET bromodomain inhibitors in triple negative breast cancer. Nature. 2015. PMID: 26735014.
20. Xu H, Xu K, He H, Zang C, Chen C, Chen Y, Qin Q, Wang S, Wang C, Hu S, Li F, Long H, Brown M, Liu X.S. Integrative Analysis Revealed the collaboration between EZH2 and E2F1 in transcriptional regulation of cancer-related genes. Molecular Cancer Research. 2015. PMID: 26659825.
21. Hsieh CL, Botta G, Gao S, Li T, Van Allen EM, Treacy DJ, Cai C, He H, Sweeney CJ, Brown M, Balk SP, Nelson PS, Garraway LA, Kantoff PW. PLZF, a Tumor Suppressor Genetically Lost in Metastatic Castration-Resistant Prostate Cancer, Is a Mediator of Resistance to Androgen Deprivation Therapy. Cancer Res. 2015. PMID: 25808865.
22. Cai C*, He H*, Chen S, Yu Z, Gao Y, Chen S, Schüle R, Liu X.S, Brown M and Balk S.P. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Reports. 2014. PMID: 25482560.
23. He H*, Meyer CA*, Hu SS, Chen MW, Zang CZ, Liu Y, Rao PK, Fei T, Xu H, Long H, Liu XS, Brown M. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nature Methods. 2014. PMID: 24317252.
24. Sun T, Wang X, He H, Sweeney C.J, Liu X.S, Brown M, Lee G.M, Kantoff P.W. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene. 2014. PMID: 23770851.
25. Yang Y, Liu R, Qiu R, Zheng Y, Huang W, Hu H, Ji Q, He H, Shang Y, Gong Y, Wang Y. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene. 2013. PMID: 24292684.
26. Jeselsohn RM, Werner L, Regan MM, Fatima A, Gilmore L, Collins LC, Beck AH, Bailey ST, He H, Buchwalter G, Brown M, Iglehart JD, Richardson A, Come SE. Digital quantification of gene expression in sequential breast cancer biopsies reveals activation of an immune response. 2013. PLoS One. PMID: 23741308.
27. May T, Yang J, Shoni M, Liu S, He H, Gali R, Ng S.K, Crum C, Berkowitz R.S and Ng S.W. BRCA1 Expression is Epigenetically Repressed in Sporadic Ovarian Cancer Cells by Overexpression of C-Terminal Binding Protein-2. Neoplasia. 2013. PMID: 23730208.
28. Cai C, Wang H, He H, Chen S, Mucci L, Wang Q, Flore C, Sowalsky A, Loda M, Liu XS, Brown M, Balk S, Yuan X. ERG Activation of an Androgen Receptor Regulated Enhancer in the SOX9 Gene Mediates Tumor Development in TMPRSS2: ERG Fusion Positive Prostate Cancer. J Clin Invest. 2013. PMID: 23426182.
29. He H*, Meyer CA*, Chen MW, Jordan VC, Brown M, Liu XS. Differential DNase I Hypersensitivity reveals Factor-dependent Chromatin Dynamics. Genome Research. 2012. PMID: 22508765.
30. Xu K, Wu Z, Groner A, He H, Cai C, Liu T, Thornton J, Gregory R, Loda M, Stack E, Kantoff P, Balk S, Liu X.S, Brown M. EZH2 Oncogenic Activity in Castration Resistant Prostate Cancer is Polycomb-Independent. Science. 2012. PMID: 23239736.
31. Xu Y, Xu C, Kato A, Tempel W, Abreu JC, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, Diao J, Wu F, He H, Cui Q, Clark E, Ma C, Barbara A, Veenstra J, Xu G, Kaiser G, Liu XS, Sugrue S, He X, Min J, Kato Y, Shi Y. Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development. Cell. 2012. PMID: 23217707.
32. Chen Y, Negre N, Li Q, Mieczkowska J, Slattery M, Liu T, Zhang Y, Kim T, He H, Zieba J, Ruan Y, Bickel P, Myers R,Wold B, White K, Lieb J, Liu X.S. Systematic evaluation of factors influencing ChIP-seq fidelity using ultra-deep sequencing. Nature Methods. 2012. PMID: 22522655.
33. Cai C, He H, Chen S, Coleman I, Wang H, Fang Z, Nelson PS, Chen S, Brown M, Balk SP. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011. PMID: 22014572.
34. Meyer CA, He H, Brown M, Liu XS. BINOCh: Binding Inference from Nucleosome Occupancy Changes. Bioinformatics. 2011. PMID: 21551136.
35. Wang Y, Chen J, Wei G, He H, Zhu X, Xiao T, Wei G, Dong B, He S, Skogerbø G, Chen R. The Caenorhabditis elegans intermediate-size transcriptome shows high degree of stage specific expression. Nucleic acids research. 2011. PMID: 21378118. IF=8.81.
36. Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He H, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder R, Brown M, Kato S. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011. PMID: 22121020.
37. He H*, Meyer CA*, Shin H, Bailey S, Wei G, Wang Q, Zhang Y, Xu K, Brown M, Liu. XS. Nucleosome Dynamics Define Transcriptional Enhancers. Nature Genetics. 2010 Apr; 42(4): 343-7. PMID: 20208536.
38. Verzi M, Shin H, He H, Sulahian R, Meyer C, Montgomery R, Fleet J, Brown M, Liu XS, Shivdasani R. Differentiation-specific histone modifications reveal dynamic chromatin interactions and alternative partners for the intestinal transcription factor CDX2. Developmental Cell. November 2010. PMID: 21074721.
39. Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, He H, Brown M, Liu XS, Davis M, Caswell JL, Beckwith CA, Hills A, Macconaill L, Coetzee GA, Regan MM, Freedman ML. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A. 2010 May 7. PMID: 20453196.
40. Li T*, He H*, Wang Y, Skogerbø G, Chen R. “In vivo analysis of Caenorhabditis elegans noncoding RNA promoter motifs”. BMC Mol Biol. 2008; 9:71. PMID: 18680611.
41. He S, Su H, Liu C, Skogerbo G, He H, He D, Zhu X, Liu T, Zhao Y, Chen R. “MicroRNA-encoding long non-coding”. BMC Genomics. 2008; 9:236. PMID: 18492288.
42. Aftab MN*, He H*, Li T, Skogerbø G, Chen R. “Microarrray Analysis of ncRNA expression patterns after RNAi against snoRNA associated proteins”. BMC Genomics. 2008; 9:278. PMID: 18547420.
43. Jia D*, Cai L*, He H*, Skogerbø G, Li T, Aftab MN, Chen R. “Systematic identification of non-coding RNA 2,2,7-trimethylguanosine cap structures in Caenorhabditis elegans”. BMC Mol Biol. 2007; 8:86. PMID: 17903271.
44. He H*, Wang J*, Liu T*, Liu XS, Li T, Wang Y, Qian Z, Zheng H, Zhu X, Wu T, Shi B, Deng W, Zhou W, Skogerbø G, Chen R. “Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray”. Genome Res. 2007; 17:1471–1477. PMID: 17785534.
45. He H*, Cai L*, Skogerbø G, Deng W, Liu T, Zhu X, Wang Y, Jia D, Zhang Z, Tao Y, Zeng H, Aftab MN, Cui Y, Liu G, Chen R. “Profiling Caenorhabditis elegans non-coding RNA expression with a combined microarray”. Nucleic Acids Res. 2006; 34: 2976–2983. PMID: 16738136.
46. Deng W, Zhu X, Skogerbø G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, Liu C, Li B, Bai B, Wang J, Jia D, Sun S, He H, Cui Y, Wang Y, Bu D, Chen R. Organization of the Caenorhabditis elegans small non-coding transcriptome: Genomic features, biogenesis, and expression. Genome Res. 2006; 16(1):20-29. PMID: 16344563.
47. Wang H, He H, Bao J. Average cycle period in asymmetrical flashing ratchet, Commun. Theor. Phys. 2005. 43-229.
.png)