费腾 博士/教授(东北大学生命科学与健康学院)
报告主题:RNA结合蛋白与环状RNA调控

嘉宾简介:
清华大学博士,哈佛大学医学院博士后,入选第十二批国家千人计划青年项目。其课题组主要运用多种高通量的功能基因组学结合精细的分子细胞学研究手段,探讨肿瘤、代谢性疾病以及干细胞领域相关的重大问题。在Cell Stem Cell, Cancer Cell, Nature Genetics, Nature Methods, Nat Struct Mol Biol. 以及PNAS等国际主流刊物上发表高水平论文多篇。
代表性成果介绍:
PNAS:前列腺癌中HNRNPL介导circRNA的形成
2017年6月13日,PNAS杂志在线发表了一篇前列腺癌中筛选RNA拼接因子的文章,介绍基于CRISPR基因打靶文库筛选体系,鉴定到与前列腺癌有关的RNA拼接因子HNRNPL,该因子也参与调控了circRNA的形成过程。文章的通讯作者为东北大学生命科学与健康学院“青年千人计划”费腾教授,哈佛大学公共健康学院/Dana-Farber癌症研究所X. Shirley Liu教授和Dana-Farber癌症研究所/哈佛大学医学院Myles Brown教授。费腾教授也是本文的共同第一作者
本文作者利用CRISPR的全基因组打靶文库筛选与前列腺癌有关的基因,聚类分析后找出了其中与RNA编辑有关的基因,其中HNRNPL是影响最为显著的。接下来作者分析了HNRNPL结合RNA的序列特异性,并基于RIP-Seq分析了该基因在前列腺癌细胞中调控RNA可变剪切和环形RNA形成的作用。(参考文献[4])
推荐阅读:PNAS报道前列腺癌相关的RNA拼接因子参与调控circRNA形成
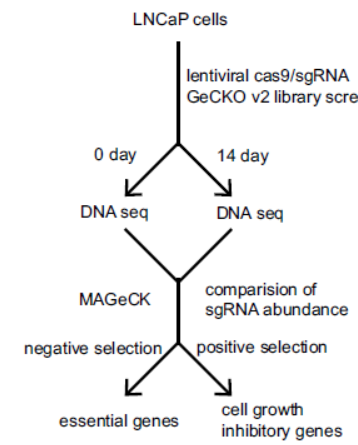
CRISPR打靶文库筛选前列腺癌关键基因的技术路线(参考文献[4])
Nature Structural & Molecular Biology:人类肿瘤相关lncRNA分析
2013年6月2日,Nature Structural & Molecular Biology以Resource形式发表了一项肿瘤相关lncRNA的研究工作,文章的通讯作者是Dana-Farber癌症研究所X. Shirley Liu教授,Myles Brown教授和Yiwen Chen教授。费腾博士是本文的共同第一作者。
本文分析了四种不同肿瘤类型中大约1300个肿瘤标本的10207个lncRNA基因的表达谱。结合临床分析的一些结果,系统分析了这些lncRNA与肿瘤亚型和临床预后的关系,预测了其中可能作为驱动因素的lncRNA。(参考文献[13])
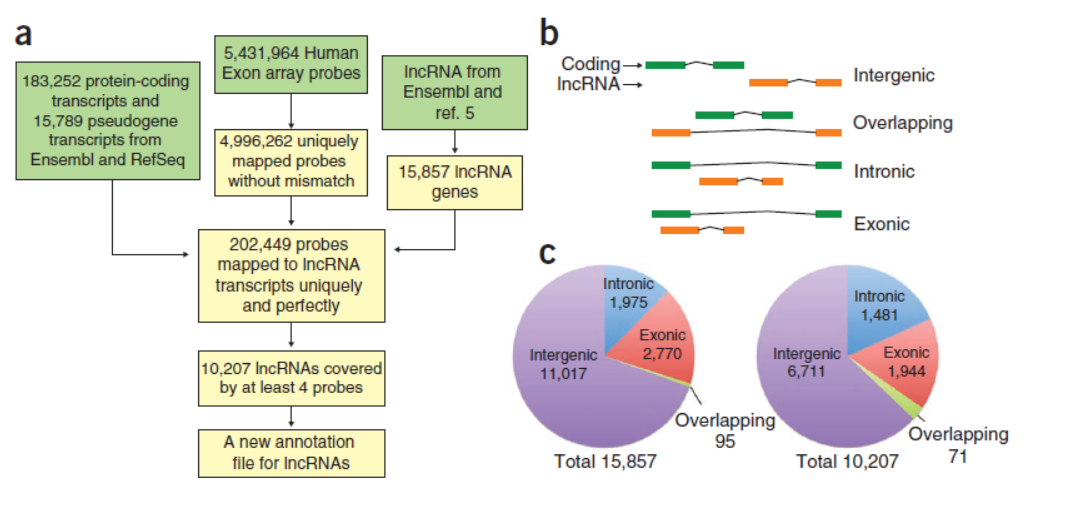
肿瘤非编码RNA分析(参考文献[13])
Cell Stem Cell:小鼠胚胎干细胞中BMP4通过DSP9调控ERK活性
2012年2月3日,Cell Stem Cell杂志发表了清华大学陈烨光教授为通讯作者的研究论文,报道发现小鼠胚胎干细胞中BMP4通过DSP9调控ERK活性。费腾博士是本文的共同第一作者。(参考文献[16])
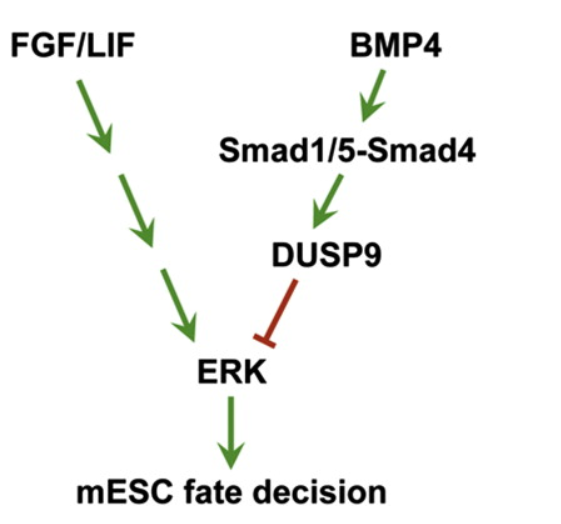
小鼠胚胎干细胞中BMP4通过DSP9调控ERK活性(参考文献[16])
Cell Research:Smad2在小鼠胚胎干细胞的中内胚层分化中介导Activin/Nodal通路
2010年11月16日,Cell Research杂志发表了清华大学陈烨光教授和中科院遗传发育所韩敬东教授为共同通讯作者的文章,报道Smad2在小鼠胚胎干细胞的中内胚层分化中介导Activin/Nodal通路。费腾博士是本文的第一作者。
本文发表之前已经报道人类Activin/Nodal通路参与调控人类胚胎干细胞的多能性,但小鼠中该通路是否也有相似的功能还未知晓。本文中作者发现Smad2介导Activin/Nodal的通路激活对小鼠胚胎干细胞的自我更新作用不大,但对准确进行内胚层的分化比较关键。ChIP-seq 分析了Smad2相关的下游基因,并进行了相关的通路分析。其中发现Tapbp 在内胚层分化中期关键作用。(参考文献[18])
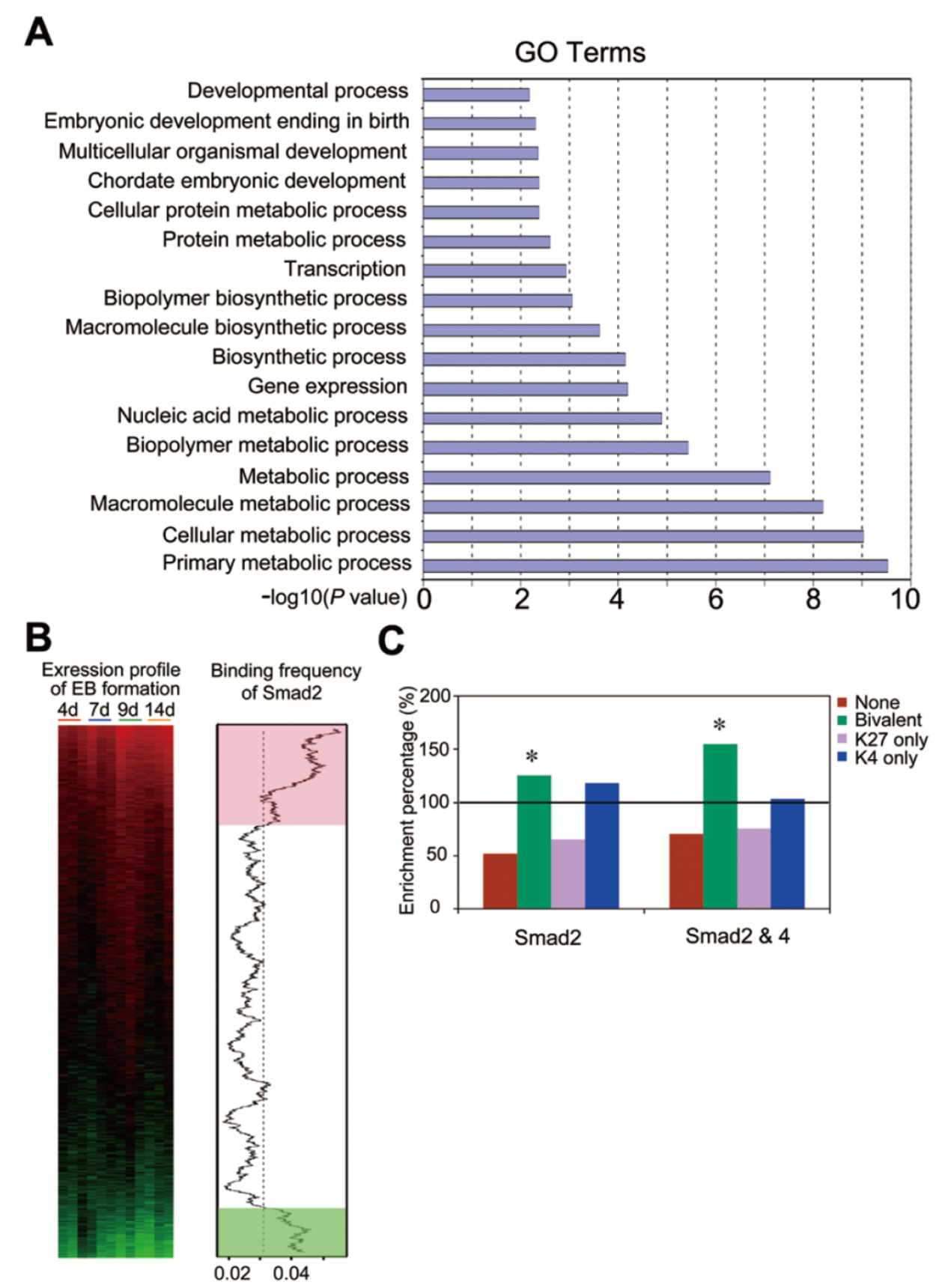
ChIP-seq 分析Smad2相关的下游基因(参考文献[18])
代表性研究成果列表
1. Jeselsohn R, Bergholz JS, Pun M, Cornwell M, Liu W, Nardone A, Xiao T, Li W, Qiu X, Buchwalter G, Feiglin A, Abell-Hart K, Fei T, Rao P, Long H, Kwiatkowski N, Zhang T, Gray N, Melchers D, Houtman R, Liu XS, Cohen O, Wagle N, Winer EP, Zhao J, Brown M. Allele-Specific Chromatin Recruitment and Therapeutic Vulnerabilities of ESR1 Activating Mutations. Cancer Cell. 2018 Feb 12;33(2):173-186.e5.
2. Willis S, Sun Y, Abramovitz M, Fei T, Young B, Lin X, Ni M, Achua J, Regan MM, Gray KP, Gray R, Wang V, Long B, Kammler R, Sparano JA, Williams C, Goldstein LJ, Salgado R, Loi S, Pruneri G, Viale G, Brown M, Leyland-Jones B. High Expression of FGD3, a Putative Regulator of Cell Morphology and Motility, Is Prognostic of Favorable Outcome in Multiple Cancers. JCO Precision Oncology. 2017 :1, 1-13. DOI: https://doi.org/10.1200/PO.17.00009
3. Fei T#. Targeting RNA binding protein in prostate cancer. Mol Cell Oncol. 2017 Jul 17;4(5):e1353855. doi: 10.1080/23723556.2017.1353855. eCollection 2017. PMID: 29057311
4. Fei T*#, Chen Y*, Xiao T, Li W, Cato L, Zhang P, Cotter MB, Bowden M, Lis RT, Zhang SG, Wu Q, Feng FY, Loda M, He HH, Liu XS#, Brown M#. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci U S A. 2017 Jun 27;114(26):E5207-E5215. doi: 10.1073/pnas.1617467114. Epub 2017 Jun 13. PMID: 28611215
5. Guo H, Ahmed M, Zhang F, Yao CQ, Li S, Liang Y, Hua J, Soares F, Sun Y, Langstein J, Li Y, Poon C, Bailey SD, Desai K, Fei T, Li Q, Sendorek DH, Fraser M, Prensner JR, Pugh TJ, Pomerantz M, Bristow RG, Lupien M, Feng FY, Boutros PC, Freedman ML, Walsh MJ, He HH. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat Genet. 2016 Oct;48(10):1142-50. doi: 10.1038/ng.3637. PMID: 27526323
6. Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW, Liu XS. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016 Mar 15;7:10982. doi: 10.1038/ncomms10982. PMID: 26975529
7. Liu Y, Fei T, Zheng X, Brown M, Zhang P, Liu XS, Wang H. An Integrative Pharmacogenomic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine. Sci Rep. 2016 Feb 26; 6:22120. doi: 10.1038/ srep22120. PMID: 26916442
8. Zhu G, Fei T, Li Z, Yan X, Chen YG. Activin Regulates Self-renewal and Differentiation of Trophoblast Stem Cells by Down-regulating the X Chromosome Gene Bcor. J Biol Chem. 2015 Sep 4;290(36):22019-29. PMID: 26221038
9. Wang H, Zheng X, Fei T, Wang J, Li X, Liu Y, Zhang F. Towards pathway-centric cancer therapies via pharmacogenomic profiling analysis of ERK signaling pathway. Clin Transl Med. 2015 Dec;4(1):66. PMID: 26220863
10. Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee GS, Chen S, Balk SP, Liu XS, Brown M, Kantoff PW. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014 May 20;111(20):7319-24. PMID: 24778216
11. He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, Liu XS, Brown M. Refined DNase seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods. 2014 Jan;11(1):73-8. PMID: 24317252
12. Wang H, Meyer CA, Fei T, Wang G, Zhang F, Liu XS. A systematic approach identifies FOXA1 as a key factor in the loss of epithelial traits during the epithelial-to-mesenchymal transition in lung cancer. BMC Genomics. 2013 Oct 4;14(1):680. PMID: 24093963
13. Du Z*, Fei T*, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y*, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013 Jul;20(7):908-13. PMID: 23728290
14. Ni M, Chen Y, Fei T, Li D, Lim E, Liu XS, Brown M. Amplitude modulation of androgen signaling by c-MYC. Genes Dev. 2013 Apr 1;27(7):734-48. PMID: 23530127
15. Zhang J*, Fei T*, Li Z*, Zhu G, Wang L, Chen YG. BMP induces cochlin expression to facilitate self-renewal and suppress neural differentiation of mouse embryonic stem cells. J Biol Chem. 2013 Mar 22;288(12):8053-60. PMID: 23344953
16. Li Z*, Fei T*, Zhang J, Zhu G, Wang L, Lu D, Chi X, Teng Y, Hou N, Yang X, Zhang H, Han JD, Chen YG. BMP4 Signaling Acts via dual-specificity phosphatase 9 to control ERK activity in mouse embryonic stem cells. Cell Stem Cell. 2012 Feb 3;10(2):171-82. PMID: 22305567
17. Sun Y*, Fei T*, Yang T*, Zhang F, Chen YG, Li H, Xu Z. The suppression of CRMP2 expression by BMP-SMAD gradient signaling controls multiple stages of neuronal development. J Biol Chem. 2010 Dec 10; 285(50):39039-50. PMID: 20926379
18. Fei T, Zhu S, Xia K, Zhang J, Li Z, Han JD, Chen YG. Smad2 mediates Activin/Nodal signaling in mesendoderm differentiation of mouse embryonic stem cells. Cell Res. 2010 Dec; 20(12):1306-18. PMID: 21079647
19. Fei T, Chen YG. Regulation of embryonic sem cell self-renewal and differentiation by TGF-β family signaling. Sci China Life Sci. 2010 Apr; 53(4):497-503. PMID: 20596917
20. Fei T*, Xia K*, Li Z, Zhou B, Zhu S, Chen H, Zhang J, Xiao H, Han J, Chen YG. Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination. Genome Res. 2010 Jan; 20(1):36 44. PMID: 19926752
21. Zhang S, Fei T, Zhang L, Zhang R, Chen F, Ning Y, Han Y, Feng XH, Meng A, Chen YG. Smad7 antagonizes transforming growth factor beta signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol Cell Biol. 2007 Jun; 27(12):4488-99. PMID: 17438144
22. Huang W, Chang HY, Fei T, Wu H, Chen YG. GSK3 beta mediates suppression of cyclin D2 expression by tumor suppressor PTEN. Oncogene. 2007 Apr 12; 26(17):2471-82. PMID: 17043650
23. Ma J, Wang Q, Fei T, Han JD, Chen YG. MCP-1 mediates TGF-beta-induced angiogenesis by stimulating vascular smooth muscle cell migration. Blood. 2007 Feb 1; 109(3):987-94. PMID: 17032917
.png)
