吴缅 博士/教授(河南省人民医院转化医学研究中心,郑州大学医学科学院)
报告主题:CircACC1在代谢应激时调控AMPK的组装和激活机制

嘉宾简介:
中国科学技术大学二级教授。1982年毕业于南京师范大学生物系;1988年获美国哥伦比亚大学分子生物学博士;1989-1991在美国哈佛大学细胞与发育生物学系做博士后。1992开始先后任新加坡国立大学和南洋理工大学助理教授和副教授。2000年入选中科院“百人计划”特聘教授。在生命科学领域的权威杂志包括:Nature Cell Biology,Molecular Cell, Cell Metabolism, PNAS, EMBO J,Nature Communications等杂志上发表论文60多篇,被引用超过3200次。目前是Acta Biochimica et Biophysica Sinica、Frontiers in Cell and Developmental Biology及中国生物化学与分子生物学报编委并担任Journal of Molecular and Cell Biology副主编。
肿瘤细胞的代谢异常是肿瘤的重要特征之一,AMPK作为细胞中重要的能量感受器,监控整个细胞的能量代谢水平并调控细胞的合成与分解代谢。AMPK本身的活性也受到严格的调控。非编码RNA作为一个新兴的功能性分子,越来越引起人们的重视,长链非编码RNA参与调控AMPK已有报道,但是环形RNA是否参与调控AMPK却未被发现。ACC1(乙酰辅酶A羧化酶,ACACA)是脂肪酸从头合成中反应中第一步的关键酶。本研究发现:血清饥饿条件会激活JNK-c-JUN,而转录因子c-JUN的激活使得ACC1基因更多表达环状RNA分子CircACC1而非线性ACC1 mRNA。CircACC1通过结合AMPK的β1和γ1调节亚基,进而稳定并激活AMPK复合物。AMPK的激活促进细胞中的脂肪酸β氧化和糖酵解。此外,在结直肠癌临床病例中,CircACC1的表达与AMPK的激活呈现正相关。因此,该研究发现了营养缺乏条件下的一条JNK-c-JUN-circACC1-AMPK细胞存活通路。通过这一信号通路,丰富了我们对环形RNA参与细胞代谢调控的认识。
代表性成果介绍:
Cell Metabolism:circACC1在代谢应激时调控AMPK的组装和激活
2019年5月30日,Cell Metabolism杂志发表的一篇研究论文,报道发现circACC1在代谢应激时调控AMPK的组装和激活。文章的通讯作者是中国科学技术大学的吴缅教授和安徽医科大学的胡汪来教授。
本文先从筛选脂代谢相关基因来源的circRNA是否与脂代谢相关入手,找到了circACC1分子,然后进一步确认了circACC1与脂代谢的相关性,进一步分析了ACC1,AMPK等代谢酶和调控基因的表达和磷酸化状态和表达水平,发现了干扰circACC1能够降低ACC1,FPKFB3的磷酸化水平,同时也降低AMPK磷酸化水平及亚基的丰度。这些发现暗示了circACC1可能与AMPK的功能有关系,相互作用分析确认了circACC1与AMPK的β和γ亚基可直接相互作用,突变分析找出了它们相互作用的区段。基于蛋白稳定性的分析发现了circACC1与AMPK的相互作用能够促进AMPK的稳定性,也同时参与维持AMPK基础活性。作者还发现了血清饥饿可通过c-Jun激活circACC1的表达。最后肿瘤学功能分析表明circACC1具有促进肿瘤生长的作用,病人标本中也存在较高比例的circACC1高表达情况。(参考文献[1])
推荐阅读:这个circRNA为什么能发表20分的文章?
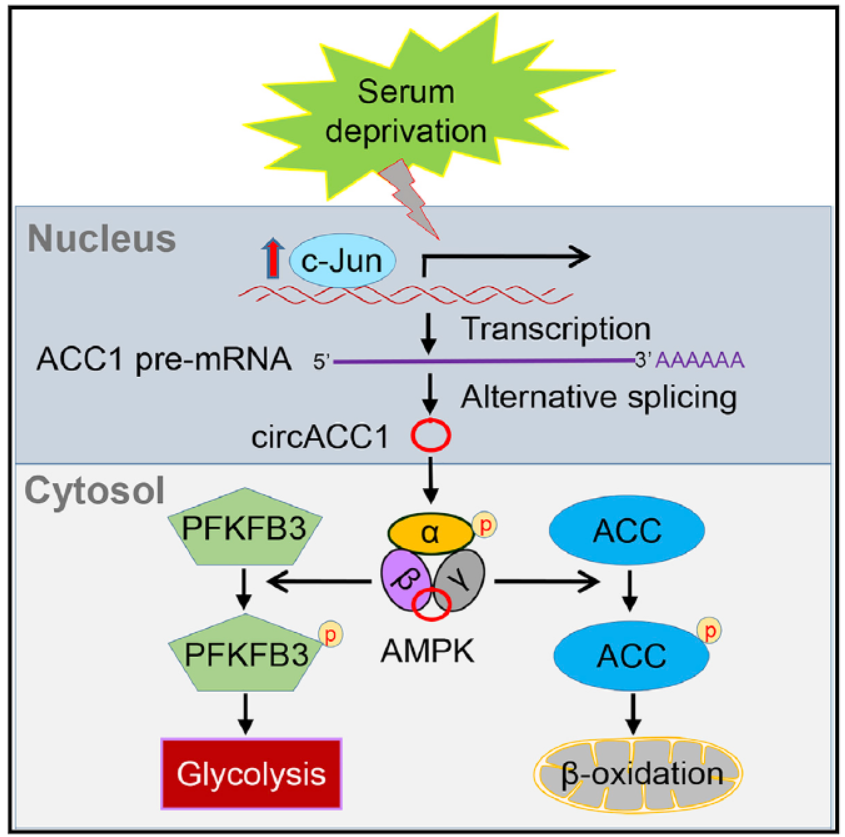
circACC1在代谢应激时调控AMPK的组装和激活(参考文献[1])
Nature Communications: 巨噬细胞中非编码RNA Neat1调控炎症小体激活
2019年4月3日,Nature Communications在线发表了中国科学技术大学吴缅教授的一篇非编码RNA的研究工作,报道发现非编码RNA Neat1在巨噬细胞中可参与炎症小体的激活。宾夕法尼亚大学杨小鲁教授是本文的共同通讯作者。
作者通过紫外交联捕获的方法,通过NLRP3的抗体捕获了相互作用的分子,基于高通量测序分析得到了若干非编码RNA分子,其中就有Neat1。功能分析发现干扰Neat1可显著降低LPS诱导的NLRP3的激活,表现为抑制Caspase-1的激活和IL-1β生成量下降。除了NLRP3,作者发现Neat1还可以激活NLRC4 和AIM2。正常状态下,Neat1处于细胞核中的paraspeckles结构中。当遇到炎症刺激信号后,Neat1释放至细胞质中,促进炎症小体(包括NLRP3,NLRC4 或AIM2等等)与Caspase-1的结合,激活Caspase-1,促进下游IL-1β等的生成,诱发炎症反应。在Neat1敲除的小鼠中,炎症刺激信号诱发的验证效应显著下降,也证明了上述通路。除了炎症刺激信号,低氧也可以诱导Neat1的表达,HIF-2α介导Neat1的表达。(参考文献[2])
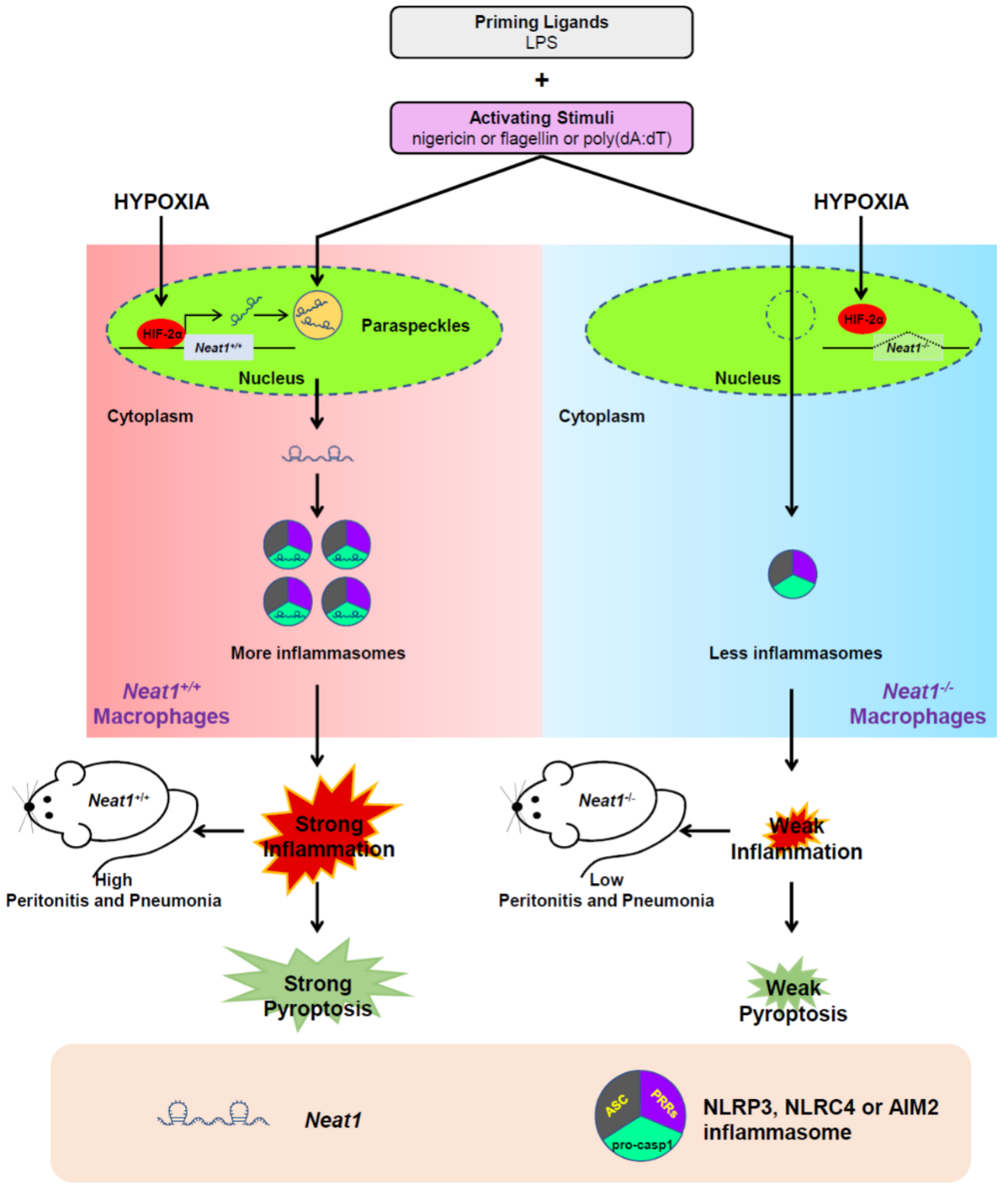
巨噬细胞中非编码RNA Neat1调控炎症小体激活(参考文献[2])
PNAS: 非编码RNA OVAAL通过双重机制促进癌细胞存活
2018年11月26日,中国科学技术大学吴缅教授与澳大利亚纽卡斯尔大学金雷教授为共同通讯作者在PNAS杂志发表了一项非编码RNA的重要工作,报道非编码RNA OVAAL通过控制RAF/MEK/ERK信号通路和p27介导的细胞衰老的双重机制促进癌细胞的存活。
TRAIL和 Mcl-1 抑制剂 UMI-77分别通过外源和内源途径诱导细胞凋亡,癌症发生过程中往往伴随凋亡通路的部分失活,分别分析可耐受两种药物处理条件的细胞中表达变化的分子有助于找到对该途径有作用的功能分子。作者首先从两种凋亡诱导条件耐受的细胞中测序分析了表达谱的变化,两种细胞中共同上调的非编码RNA分子有37种,其中OVAAL (ovarian adenocarcinoma-amplified lncRNA)发现在多种肿瘤细胞抗凋亡机制中发挥作用。机制分析表明OVAAL可以一方面通过结合STK3,促进下游RAF级联通路,最终促进c-Myc和Mcl-1表达,促进增殖并抑制凋亡。另一方面,OVAAL还可以通过结合PTBP1,竞争性释放p27 mRNA,进而促进p27的蛋白表达,抑制细胞衰老。(参考文献[3])
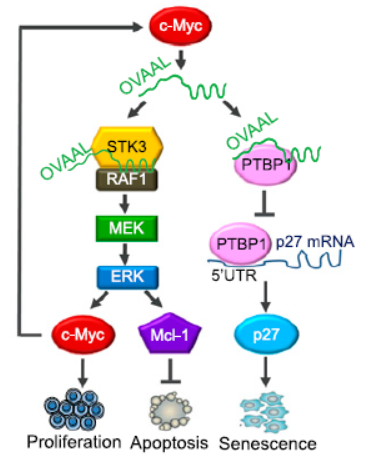
OVAAL通过双重机制促进癌细胞存活(参考文献[3])
Nature Cell Biology: 非编码RNA GUARDIN调控基因组稳定性
2018年3月28日,Nature Cell Biology杂志发表了一项非编码RNA的研究工作,发现一种受p53调控的非编码RNA GUARDIN参与了基因组稳定性的调控,中国科大吴缅教授和澳大利亚纽卡斯尔大学张旭东教授是本文的共同通讯作者。
文章作者发现GUARDIN通过两种途径参与基因组稳定性的维持,(1)竞争性结合miR-23a,促进TRF2的稳定性,进而维持端粒的稳定性。(2)GUARDIN作为支架分子,促进BRCA1与BARD1的结合,维持DNA的修复功能。干扰GUARDIN可诱发细胞凋亡或衰老。(参考文献[5])
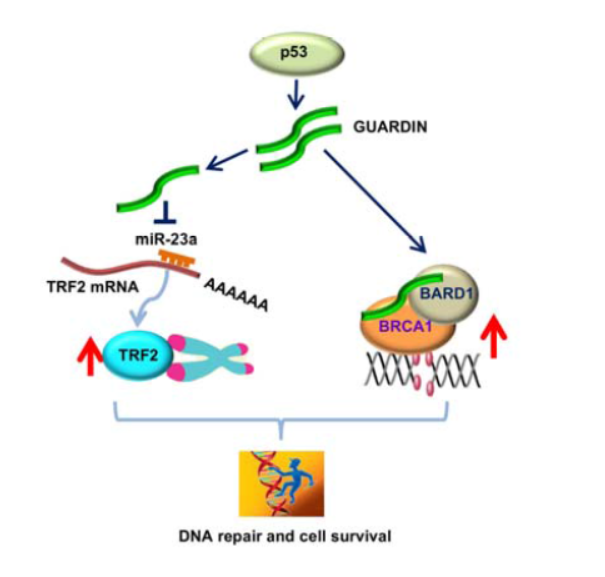
GUARDIN调控基因组稳定性(参考文献[5])
PNAS:lncRNA IDH1-AS1衔接c-Myc和HIF1α的功能
2018年1月29日,中国科技大学吴缅教授与澳大利亚纽卡斯尔大学张旭东教授为共同通讯作者,在PNAS在线发表了一项非编码RNA的研究工作,报道IDH1-AS1通过DH1介导的Warburg效应衔接c-Myc和HIF1α的功能。
有氧糖酵解(也称为Warburg效应)是肿瘤中重要的糖代谢方式,常氧条件下c-Myc是调控Warburg效应的重要因子,而在低氧条件下HIF1α诱导表达并调控糖酵解作用。有趣的是,不少研究发现常氧条件下HIF1α也表现出较高的表达水平。本文中作者发现常氧条件下c-Myc抑制非编码RNA IDH1-AS1的表达。IDH1-AS1可结合IDH1并促进其二聚化,进而促进IDH1的活性。因此在常氧的条件下,c-Myc通过抑制IDH1-AS1的表达,削弱了IDH1的活性,促进α-酮戊二酸(α-KG)的下降和ROS的增高,两者协同作用,促进HIF1α的表达升高。(参考文献[6])
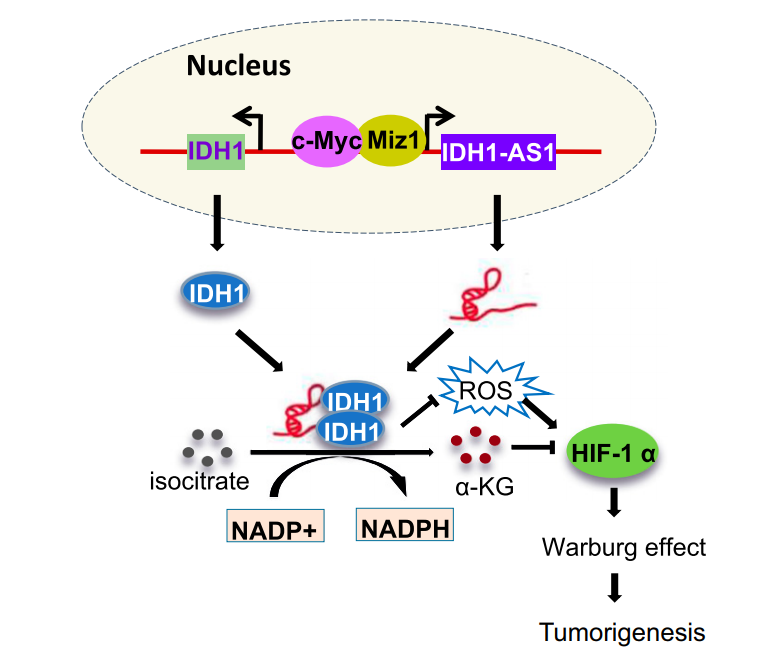
IDH1-AS1衔接c-Myc和HIF1α的功能(参考文献[6])
eLife: 非编码RNA LAST通过CNBP促进CCND1的mRNA稳定性
2017年12月4日,eLife杂志发表了中国科学技术大学吴缅教授的一篇研究论文,报道非编码RNA LAST可通过与CNBP结合促进CCND1的mRNA稳定性,促进细胞周期进程。
LAST是一种受c-Myc转录激活非编码RNA(LncRNA-Assisted Stabilization of Transcript),LAST可促进细胞周期进程,促进CCND1的mRNA稳定性。RNA Pull-down实验发现了LAST与CNBP的相互作用。体内,LAST具有促癌作用。(参考文献[9])
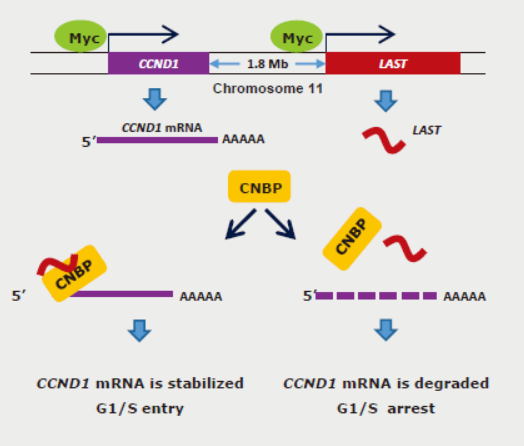
LAST通过CNBP促进CCND1的mRNA稳定性(参考文献[9])
EMBO Journal: 非编码RNA TRINGS保护细胞在葡萄糖饥饿状态下存活
2017年10月18日, EMBO Journal在线发表了中国科学技术大学吴缅教授和武汉大学宋质银教授为共同通讯作者的文章,报道受p53调控的非编码RNA TRINGS 保护细胞在葡萄糖饥饿状态下的存活。
TRINGS 是一种受p53调控的非编码RNA,在葡萄糖缺失的状态下,TRINGS 表达激活,与STRAP结合,释放GSK3β,促进GSK3β的Ser-9磷酸化,进而稳定NF-КB通路,促进细胞存活。(参考文献[10])
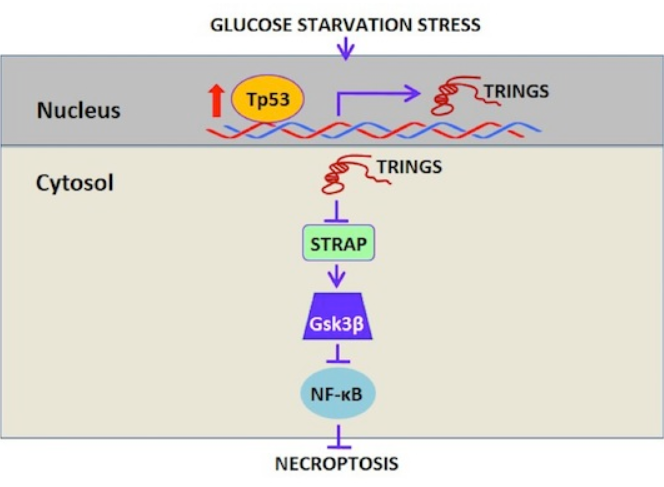
TRINGS保护细胞在葡萄糖饥饿状态下的存活(参考文献[10])
EMBO Reports:非编码RNA MIF通过竞争性结合miR-586促进FBXW7稳定
2016年6月17日,EMBO Reports杂志发表了中国科技大学吴缅教授为通讯作者的文章,报道一种受c-Myc调控的非编码RNA MIF可作为内源竞争性RNA分子(ceRNA)结合miR-586,促进FBXW7表达。(参考文献[15])
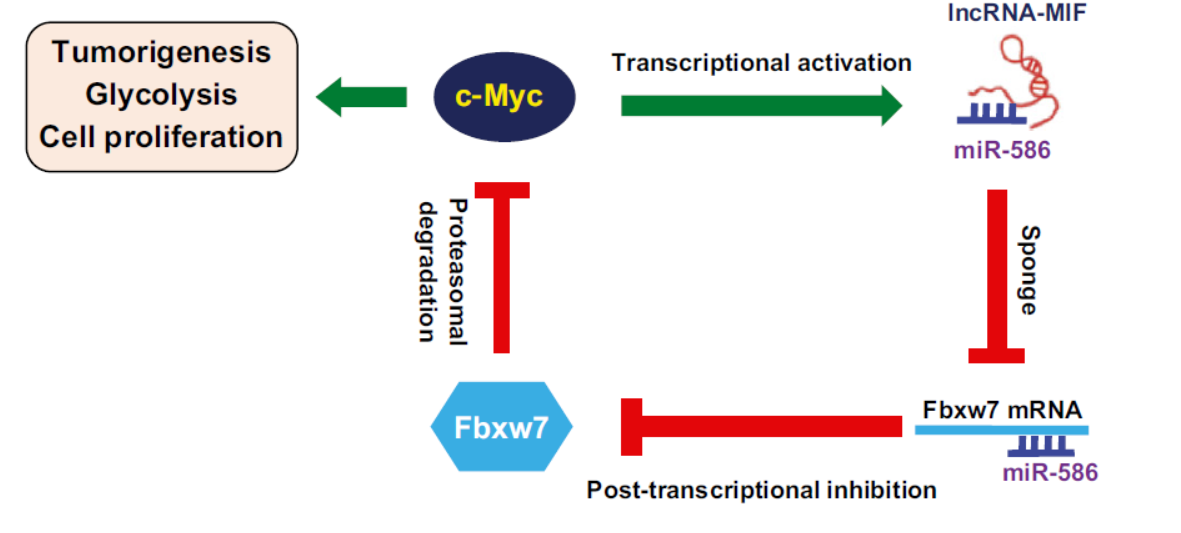
MIF竞争性结合miR-586促进FBXW7稳定(参考文献[15])
Molecular Cell: lincRNA-p21与HIF1α协同调控Warburg效应
2014年1月9日,Molecular Cell杂志发表了中国科技大学吴缅教授和梅一德教授为共同通讯作者的文章,介绍非编码RNA lincRNA-p21与HIF1α协同调控Warburg效应。
lincRNA-p21可以在低氧条件下诱导表达,也可以通过HIF1α调控表达。lincRNA-p21可以结合HIF1α或VHL,阻止VHL与HIF1α的直接结合,保护HIF1α不受VHL的降解。HIF1α又反过来诱导lincRNA-p21的表达, 形成了一种正反馈的通路。(参考文献[22])
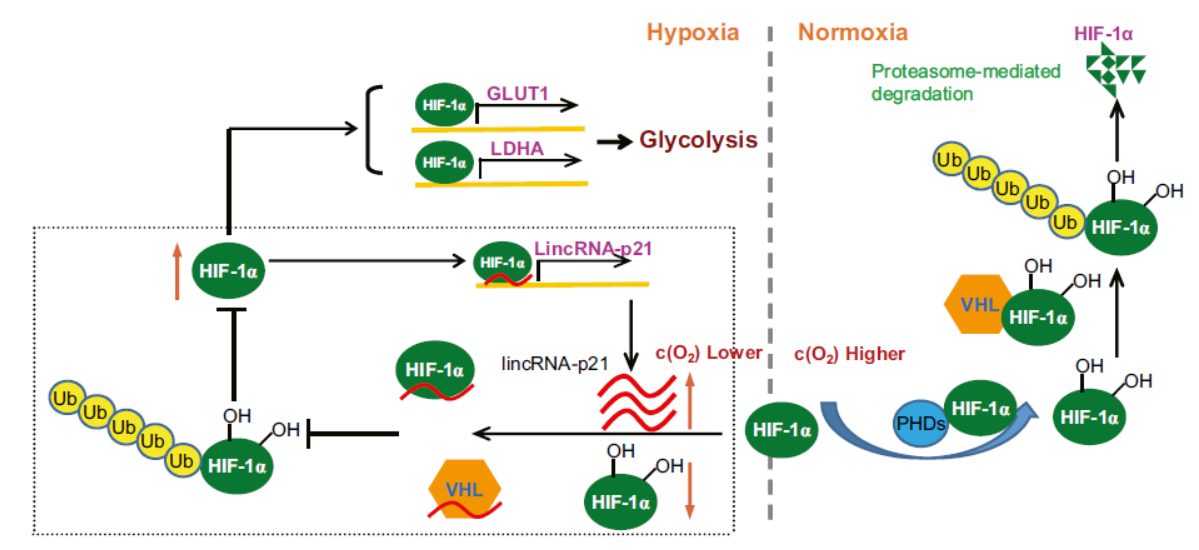
lincRNA-p21与HIF1α协同调控Warburg效应(参考文献[22])
上面列出的是吴缅教授最近发表的非编码RNA相关的工作,吴缅教授还有很多其他的高水平的研究论文,感兴趣的读者可以查阅相关的文献拜读一下。
已发文章列表:
1. Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, Thorne RF, Zhang XD, Hu W*, Wu M*. CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell Metab. 2019 May 27. pii: S1550-4131(19)30249-9. doi: 10.1016/j.cmet.2019.05.009
2. Zhang P, Cao L, Zhou R, Yang X*, Wu M*. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019 Apr 2;10(1):1495. doi: 10.1038/s41467-019-09482-6
3. Sang B, Zhang YY, Guo ST, Kong LF, Cheng Q, Liu GZ, Thorne RF, Zhang XD, Jin L*, Wu M*. Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11661-E11670. doi: 10.1073/pnas.1805950115
4. Khan MR, Wu M*, Liu G*. Tumor-suppressive or tumor-supportive: For p53, that is the question. Mol Cell Oncol. 2018 Feb 27;5(3):e1408537. doi: 10.1080/23723556.2017.1408537
5. Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD*, Wu M*. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. 2018 Apr;20(4):492-502. doi: 10.1038/s41556-018-0066-7
6. Xiang S, Gu H, Jin L, Thorne RF, Zhang XD*, Wu M*. LncRNA IDH1-AS1 links the functions of c-Myc and HIF1α via IDH1 to regulate the Warburg effect. Proc Natl Acad Sci U S A. 2018 Feb 13;115(7):E1465-E1474. doi: 10.1073/pnas.1711257115
7. Zhao K, Yang Y, Zhang G, Wang C, Wang D, Wu M*, Mei Y*. Regulation of the Mdm2-p53 pathway by the ubiquitin E3 ligase MARCH7.EMBO Rep. 2018 Feb;19(2):305-319. doi: 10.15252/embr.201744465
8. Khan MR, Bukhari I, Junxiang T, Hui L, Muhammad N, Fan C, Zhu J*, Wu M*. A Novel Sex Chromosome Mosaicism 45,X/45,Y/46,XY/46,YY/47,XYY Causing Ambiguous Genitalia. Ann Clin Lab Sci. 2017 Nov;47(6):761-764.
9. Cao L, Zhang P, Li J, Wu M*. LAST, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. Elife. 2017 Dec 4;6. pii: e30433. doi: 10.7554/eLife.30433
10. Khan MR, Xiang S, Song Z*, Wu M*. The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. EMBO J. 2017 Dec 1;36(23):3483-3500. doi: 10.15252/embj.201696239
11. Qiao M, Wu M*, Shi R*, Hu W*. PHLDA3 impedes somatic cell reprogramming by activating Akt-GSK3β pathway. Sci Rep. 2017 Jun 6;7(1):2832. doi: 10.1038/s41598-017-02982-9
12. Wang L, Zhang T, Wang L, Cai Y, Zhong X, He X, Hu L, Tian S, Wu M, Hui L, Zhang H, Gao P. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 2017 May 15;36(10):1330-1347. doi: 10.15252/embj.201695417.
13. Yang K, Wang M, Zhao Y, Sun X, Yang Y, Li X, Zhou A, Chu H, Zhou H, Xu J, Wu M, Yang J, Yi J. A redox mechanism underlying nucleolar stress sensing by nucleophosmin. Nat Commun. 2016 Nov 25;7:13599. doi: 10.1038/ncomms13599.
14. Mei Y, Wu M*. Noncoding RNAs Regulating p53 and c-Myc Signaling.Adv Exp Med Biol. 2016;927:337-65. doi: 10.1007/978-981-10-1498-7_13.
15. Zhang P, Cao L, Fan P, Mei Y, Wu M*. LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep. 2016 Aug;17(8):1204-20. doi: 10.15252/embr.201642067
16. Cheng B, Xu A, Qiao M, Wu Q, Wang W*, Mei Y*, Wu M*. BECN1s, a short splice variant of BECN1, functions in mitophagy. Autophagy. 2015 Nov 2;11(11):2048-2056. doi: 10.1080/15548627.2015.1100785.
17. Xie W, Zhang L, Jiao H, Guan L, Zha J, Li X, Wu M, Wang Z, Han J, You H. Chaperone-mediated autophagy prevents apoptosis by degrading BBC3/PUMA. Autophagy. 2015; 11(9):1623-35. doi: 10.1080/15548627.2015.1075688.
18. Gu H, Li Q, Huang S, Lu W, Cheng F, Gao P, Wang C, Miao L*, Mei Y*, Wu M*. Mitochondrial E3 ligase March5 maintains stemness of mouse ES cells via suppression of ERK signaling. Nat Commun. 2015 Jun 2;6:7112. doi: 10.1038/ncomms8112
19. Wang J, Qiao M, He Q, Shi R, Loh SJ, Stanton LW*, Wu M*. Pluripotency Activity of Nanog Requires Biochemical Stabilization by Variant Histone Protein H2A.Z. Stem Cells. 2015 Jul;33(7):2126-34. doi: 10.1002/stem.2011
20. Chu B, Wu T, Miao L*, Mei Y*, Wu M*. MiR-181a regulates lipid metabolism via IDH1. Sci Rep. 2015 Mar 5;5:8801. doi: 10.1038/srep08801
21. Chu B, Zhong L, Dou S, Wang J, Li J, Wang M, Shi Q*, Mei Y*, Wu M*. miRNA-181 regulates embryo implantation in mice through targeting leukemia inhibitory factor.J Mol Cell Biol. 2015 Feb;7(1):12-22. doi: 10.1093/jmcb/mjv006
22. Yang F, Zhang H, Mei Y*, Wu M*. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014 Jan 9;53(1):88-100. doi: 10.1016/j.molcel.2013.11.004.
23. Tang A, Mei B, Wang W, Hu W, Li F, Zhou J, Yang Q, Cui H, Wu M, Liang G. FITC-quencher based caspase 3-activatable nanoprobes for effectively sensing caspase 3 in vitro and in cells. Nanoscale. 2013 Oct 7;5(19):8963-7. doi: 10.1039/c3nr03339b
24. Du W, Jiang P, Mancuso A, Stonestrom A, Brewer MD, Minn AJ, Mak TW, Wu M*, Yang X*. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat Cell Biol. 2013 Aug;15(8):991-1000. doi: 10.1038/ncb2789
25. Huang X, Wu Z, Mei Y*, Wu M*. XIAP inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J. 2013 Aug 14;32(16):2204-16. doi: 10.1038/emboj.2013.133
26. Wang X, Zhao X, Gao P, Wu M*. c-Myc modulates microRNA processing via the transcriptional regulation of Drosha. Sci Rep. 2013;3:1942. doi: 10.1038/srep01942.
27. Yang F, Miao L, Mei Y*, Wu M*. Retinoic acid-induced HOXA5 expression is co-regulated by HuR and miR-130a. Cell Signal. 2013 Jun;25(6):1476-85. doi: 10.1016/j.cellsig.2013.03.015
28. Wang X, Zha M, Zhao X, Jiang P, Du W, Tam AY, Mei Y*, Wu M*. Siva1 inhibits p53 function by acting as an ARF E3 ubiquitin ligase. Nat Commun. 2013;4:1551. doi: 10.1038/ncomms2533.
29. Han C, Gu H, Wang J, Lu W, Mei Y*, Wu M*. Regulation of L-threonine dehydrogenase in somatic cell reprogramming. Stem Cells. 2013 May;31(5):953-65. doi: 10.1002/stem.1335.
30. Wang X, Zhao X, Gao X, Mei Y*, Wu M*. A new role of p53 in regulating lipid metabolism. J Mol Cell Biol. 2013 Apr;5(2):147-50. doi: 10.1093/jmcb/mjs064
31. Han C, Jin L, Mei Y*, Wu M*. Endoplasmic reticulum stress inhibits cell cycle progression via induction of p27 in melanoma cells. Cell Signal. 2013 Jan;25(1):144-9. doi: 10.1016/j.cellsig.2012.09.023
32. Qiu Y, Liu L, Zhao C, Han C, Li F, Zhang J, Wang Y, Li G, Mei Y, Wu M, Wu J, Shi Y. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 2012 Jun 15;26(12):1376-91. doi: 10.1101/gad.188359.112
33. Wang J, He Q, Han C, Gu H, Jin L, Li Q, Mei Y*, Wu M*. p53-facilitated miR-199a-3p regulates somatic cell reprogramming. Stem Cells. 2012 Jul;30(7):1405-13. doi: 10.1002/stem.1121.
34. Mei Y*, Wu M*.Multifaceted functions of Siva-1: more than an Indian God of Destruction. Protein Cell. 2012 Feb;3(2):117-22. doi: 10.1007/s13238-012-2018-5
35. Xie C, Wang W, Yang F, Wu M*, Mei Y*. RUVBL2 is a novel repressor of ARF transcription. FEBS Lett. 2012 Feb 17;586(4):435-41. doi: 10.1016/j.febslet.2012.01.026
36. Yi Q, Zhao X, Huang Y, Ma T, Zhang Y, Hou H, Cooke HJ, Yang DQ, Wu M, Shi Q. p53 dependent centrosome clustering prevents multipolar mitosis in tetraploid cells. PLoS One. 2011;6(11):e27304. doi: 10.1371/journal.pone.0027304
37. Jin L, Hu WL, Jiang CC, Wang JX, Han CC, Chu P, Zhang LJ, Thorne RF, Wilmott J, Scolyer RA, Hersey P, Zhang XD*, Wu M*. MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc Natl Acad Sci U S A. 2011 Sep 20;108(38):15840-5. doi: 10.1073/pnas.1019312108
38. Li N, Jiang P, Du W, Wu Z, Li C, Qiao M, Yang X*, Wu M*. Siva1 suppresses epithelial-mesenchymal transition and metastasis of tumor cells by inhibiting stathmin and stabilizing microtubules. Proc Natl Acad Sci U S A. 2011 Aug 2;108(31):12851-6. doi: 10.1073/pnas.1017372108
39. Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M*, Yang X*. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011 Mar;13(3):310-6. doi: 10.1038/ncb2172
40. Tian X, Chen Y, Hu W, Wu M*. E2F1 inhibits MDM2 expression in a p53-dependent manner. Cell Signal. 2011 Jan;23(1):193-200. doi: 10.1016/j.cellsig.2010.09.003
41. Wang G, Gao X, Huang Y, Yao Z, Shi Q, Wu M*. Nucleophosmin/B23 inhibits Eg5-mediated microtubule depolymerization by inactivating its ATPase activity. J Biol Chem. 2010 Jun 18;285(25):19060-7. doi: 10.1074/jbc.M110.100396
42. Xie W, Jin L, Mei Y, Wu M*. E2F1 represses beta-catenin/TCF activity by direct up-regulation of Siah1.J Cell Mol Med. 2009 Aug;13(8B):1719-27
43. Li M, Feng S, Wu M*. Multiple roles for nuclear localization signal (NLS, aa 442–472) of receptor interacting protein 3 (RIP3). Biochem Biophys Res Commun. 2008 Aug 8;372(4):850-5. doi: 10.1016/j.bbrc.2008.05.144
44. Mei Y, Xie C, Xie W, Tian X, Li M, Wu M*. Noxa/Mcl-1 balance regulates susceptibility of cells to camptothecin-induced apoptosis. Neoplasia. 2007 Oct;9(10):871-81.
45. Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, Castanares M, Wu M*. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal. 2007 Oct;19(10):2056-67. Epub 2007 Jun 14.
46. Duan S, Yao Z, Hou D, Wu Z, Zhu WG, Wu M*. Phosphorylation of Pirh2 by calmodulin-dependent kinase II impairs its ability to ubiquitinate p53. EMBO J. 2007 Jul 11;26(13):3062-74
47. Jiang P, Du W, Wu M*. p53 and Bad: remote strangers become close friends. Cell Res. 2007 Apr;17(4):283-5
48. Yao Z, Duan S, Hou D, Heese K, Wu M*. Death effector domain DEDa, a self-cleaved product of caspase-8/Mch5, translocates to the nucleus by binding to ERK1/2 and upregulates procaspase-8 expression via a p53-dependent mechanism. EMBO J. 2007 Feb 21;26(4):1068-80.
49. Jiang P, Du W, Heese K, Wu M*. The Bad guy cooperates with good cop p53: Bad is transcriptionally up-regulated by p53 and forms a Bad/p53 complex at the mitochondria to induce apoptosis. Mol Cell Biol. 2006 Dec;26(23):9071-82
50. Feng S, Ma L, Yang Y, Wu M*. Truncated RIP3 (tRIP3) acts upstream of FADD to induce apoptosis in the human hepatocellular carcinoma cell line QGY-7703. Biochem Biophys Res Commun. 2006 Sep 1;347(3):558-65
51. Xie W, Jiang P, Miao L, Zhao Y, Zhimin Z, Qing L, Zhu WG, Wu M*. Novel link between E2F1 and Smac/DIABLO: proapoptotic Smac/DIABLO is transcriptionally upregulated by E2F1. Nucleic Acids Res. 2006 Apr 14;34(7):2046-55
52. Mei Y, Du W, Yang Y, Wu M*. Puma(*)Mcl-1 interaction is not sufficient to prevent rapid degradation of Mcl-1. Oncogene. 2005 Nov 3;24(48):7224-37.
53. Yang Y, Hu W, Feng S, Ma J, Wu M*. RIP3 beta and RIP3 gamma, two novel splice variants of receptor-interacting protein 3 (RIP3), downregulate RIP3-induced apoptosis. Biochem Biophys Res Commun. 2005 Jun 24;332(1):181-7.
54. Song Z, Wu M*. Identification of a novel nucleolar localization signal and a degradation signal in Survivin-deltaEx3: a potential link between nucleolus and protein degradation. Oncogene. 2005 Apr 14;24(16):2723-34
55. Yang Y, Ma J, Chen Y, Wu M*. Nucleocytoplasmic shuttling of receptor-interacting protein 3 (RIP3): identification of novel nuclear export and import signals in RIP3. J Biol Chem. 2004 Sep 10;279(37):38820-9
56. Höti N, Zhu DE, Song Z, Wu Z, Tabassum S, Wu M*. p53-dependent apoptotic mechanism of a new designer bimetallic compound tri-phenyl tin benzimidazolethiol copper chloride (TPT-CuCl2): in vivo studies in Wistar rats as well as in vitro studies in human cervical cancer cells.J Pharmacol Exp Ther. 2004 Oct;311(1):22-33
57. Song Z, Liu S, He H, Hoti N, Wang Y, Feng S, Wu M*. A single amino acid change (Asp 53 –> Ala53) converts Survivin from anti-apoptotic to pro-apoptotic.Mol Biol Cell. 2004 Mar;15(3):1287-96
58. Höti N, Ma J, Tabassum S, Wang Y, Wu M*. Triphenyl tin benzimidazolethiol, a novel antitumor agent, induces mitochondrial-mediated apoptosis in human cervical cancer cells via suppression of HPV-18 encoded E6. J Biochem. 2003 Oct;134(4):521-8
59. Miao L, Yi P, Wang Y, Wu M*. Etoposide upregulates Bax-enhancing tumour necrosis factor-related apoptosis inducing ligand-mediated apoptosis in the human hepatocellular carcinoma cell line QGY-7703. Eur J Biochem. 2003 Jul;270(13):2721-31.
60. Song Z, Yao X, Wu M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J Biol Chem. 2003 Jun 20;278(25):23130-40.
61. Yi P, Zhang W, Zhai Z, Miao L, Wang Y, Wu M. Bcl-rambo beta, a special splicing variant with an insertion of an Alu-like cassette, promotes etoposide- and Taxol-induced cell death. FEBS Lett. 2003 Jan 16;534(1-3):61-8.
62. Yang Y, Ma J, Song Z, Wu M. HIV-1 TAT-mediated protein transduction and subcellular localization using novel expression vectors. FEBS Lett. 2002 Dec 4;532(1-2):36-44
.png)
